Contents
- Introduction
- My Requirements
- The Solution: Pubmed-Sieve RSS Generation
- Create a literature feed with Pubmed-Sieve
- Pubmed-Sieve Features & Workflow Tips
Introduction
Keeping up with the ever-expanding medical and life science literature is incredibly important but far from straightforward. Thousands of papers are published each day, which makes finding the signal in the noise a hard problem.
Over the course of my scientific career (in which I’ve changed fields a couple times) I’ve developed an approach and related tools that others might find helpful. This post is to summarize my methodology for following the literature in life science.
My Requirements
The following attributes are essential for me. Highlighted in red are those that were particularly hard to satisfy with existing tools.
- Feeds showing titles, abstracts, and authors that I can quickly browse on my computer or iPhone.
- Ability to create feeds that are a combination of author names (and possibly author position), keywords, journal names, etc.
- Filters that yield a volume of papers that I can keep up with every day (~10s of abstracts a day).
- High quality results – I shouldn’t be missing any papers related to my work (and getting caught by surprise).
- Ability to quickly & painlessly tweak my feeds.
- Free and ideally built on open source infrastructure (FOSS).
The Solution: Pubmed-Sieve RSS Generation
Essentially all life science literature is indexed on PubMed (even preprints now). PubMed actually supports creating RSS feeds for arbitrary searches. However, creating queries by hand is very time consuming (imagine you have numerous keywords/authors), and any modification requires manual regeneration of the RSS link.
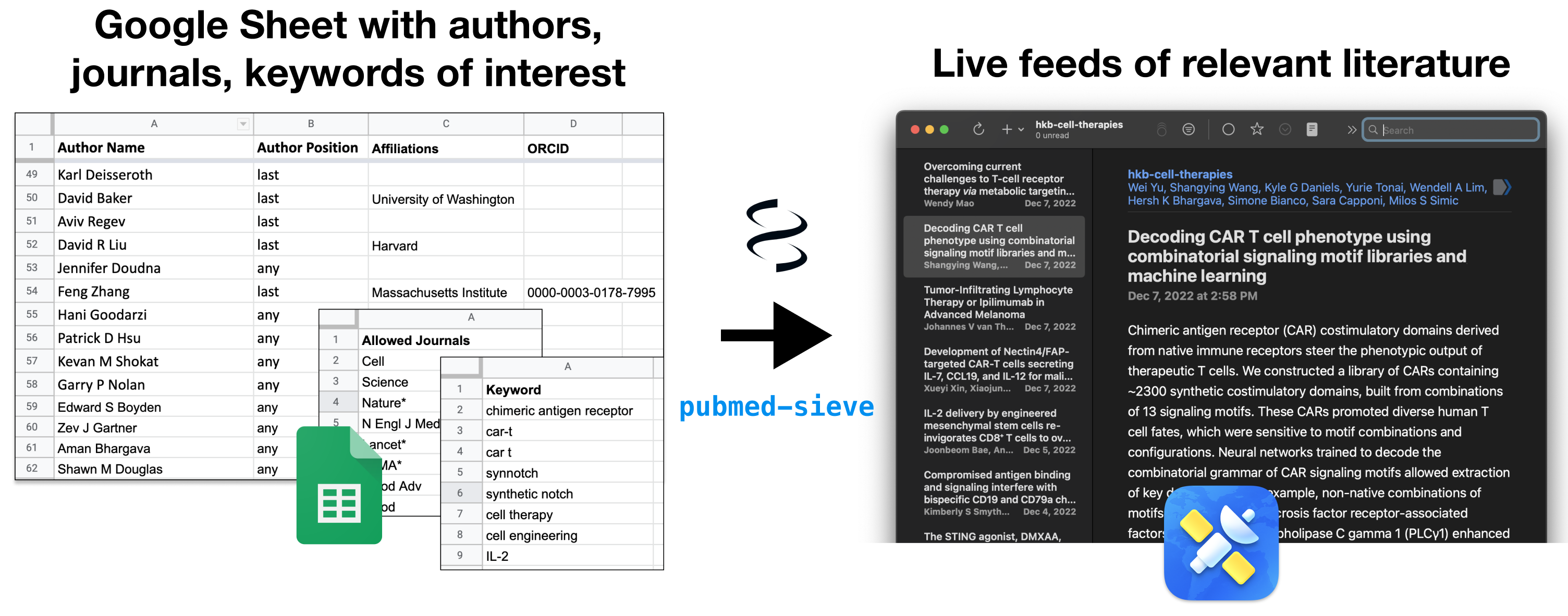
Cue a small tool I built to solve this problem: Pubmed-Sieve (GitHub). It takes a Google Sheet (example) specifying a list of authors and/or keywords and/or journals and outputs 1) a PubMed search string and 2) a PubMed RSS Feed link for that search, ready to plug into your favorite RSS reader. Pubmed-Sieve is written in Python, and can be run on the web using Binder (a free tool for running Jupyter Notebooks in the cloud: ). You can also run it on your own machine/infra if you prefer.
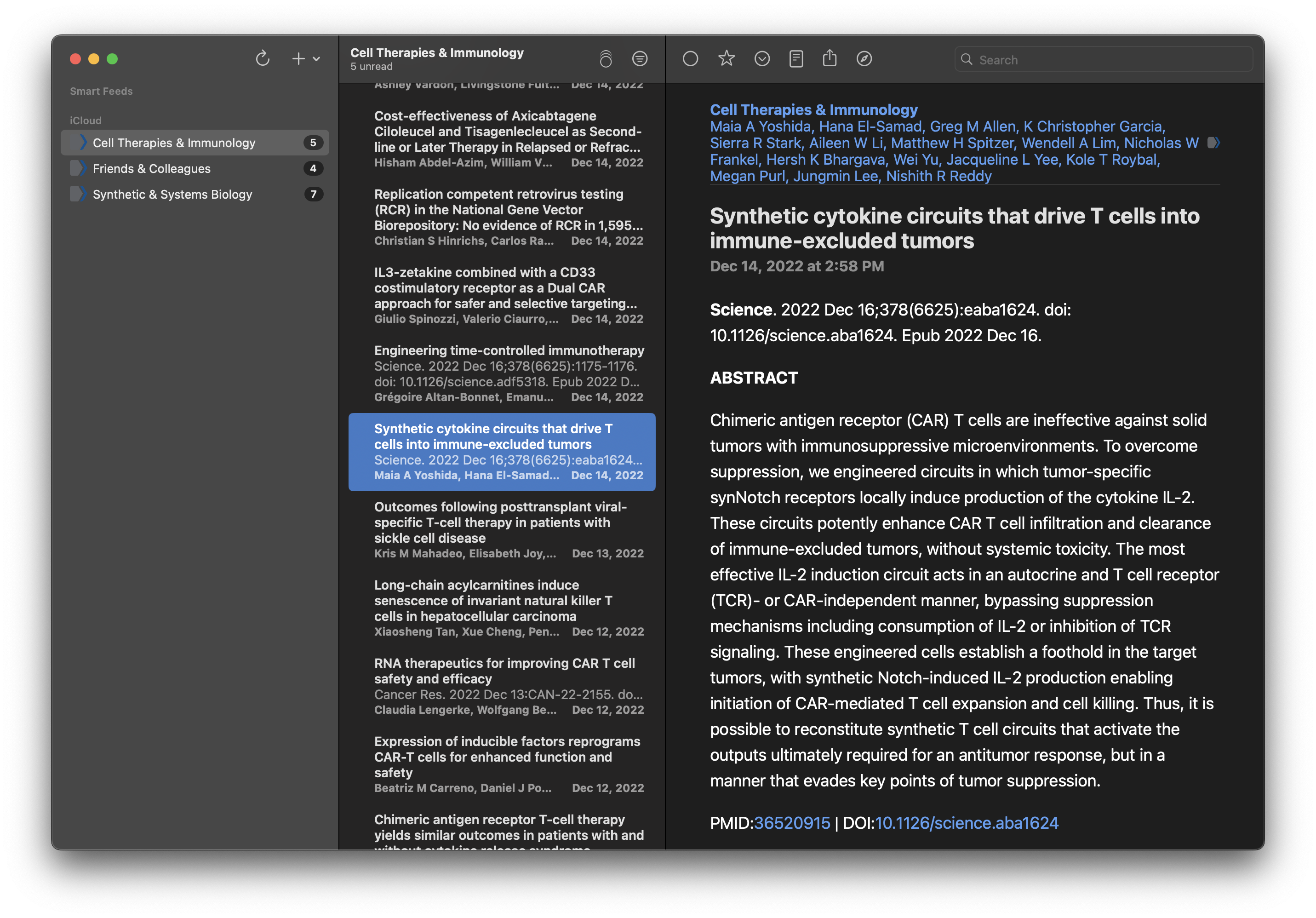
Here’s what my final feeds look like (read in NetNewsWire):
Create a literature feed with Pubmed-Sieve
-
Make a copy of this Google Sheet template (File > Make a Copy; make sure its set to ‘anyone with the link can view’) and add your authors (sheet 1) and/or keywords+journals (sheet 2) of interest. By default, Pubmed-Sieve will filter for
[Authors] OR [Keywords AND Journals]1. Here’s a link to an example spreadsheet. -
Launch Pubmed-Sieve on Binder here:
(or clone and run the Github repo).
-
Paste your spreadsheet URL and run the Notebook. A few seconds later, you’ll get your RSS feed URL2 and query string!
The output will look something like this:
-
Pubmed RSS Feed URL: https://pubmed.ncbi.nlm.nih.gov/rss/search/1rSUu–tbw4_4aTL1f48QYoTEf3gyx4zvTK4-A2V3FH_VDx1Af/?limit=100&utm_campaign=pubmed-2&fc=20230115020214
-
Raw Pubmed Query String:
(((Aviv Regev[FAU])) OR ((Carl H June[FAU] AND June CH[lastau])) OR ((Crystal L Mackall[FAU])) OR ((David Baker[FAU]) AND (University of Washington[affil])) OR ((David R Liu[FAU]) AND (Harvard[affil])) OR...
I recommend using the FOSS tool NetNewsWire (macOS, iOS) or the Freemium tool Feedly (Mac, Windows, Linux, iOS, Android) as your RSS reader. Simply paste the RSS feed URL into your reader and you’re good to go!
I maintain multiple feeds to keep things organized: one for my primary research interests, one for my secondary interests, and one for my friends + collaborators. Here’s an example of a feed spreadsheet, and the corresponding Pubmed search results.
Pubmed-Sieve Features & Workflow Tips
Rapid Iteration on Feeds: You can quickly modify your feeds by adding/removing authors/keywords/journals and re-running the notebook. This is much faster than manually modifying the PubMed search string and re-generating the RSS feed. I update my feeds whenever I meet someone new or read a paper that I think is relevant.
Advanced tip: If you don’t want to have to update the RSS link in your feed reader all the time, you can use a link shortener like bit.ly as a middleman, and simply update the short link. This could probably be automated, too.
Sophisticated Author Search Criteria: You can specify author names, positions (e.g. first/last), institution, or even ORCiD. This is great for authors with ambiguous names, or if you only want to follow an author’s main projects (some people are middle author on huge numbers of papers).
Automatically filter out non-papers: Pubmed-Sieve adds flags to eliminate non-papers (e.g. errata, news pieces, letters to the editor, etc.) from your feed.
Journal Whitelist: You can specify a list of journals that you want to allow papers from. You can even use wildcards (e.g. “Cell*” to allow all Cell family journals).
Pubmed Search Lifehacks:
[tiab]flag to stipulate that a search term should be in the title and/or abstract, e.g."CAR-T cells" [tiab].- Author position flags:
[1au]for first author,[lastau]for last author,[au]for any author. - Use the
[hasabstract]flag to only return papers with abstracts, which is great for filtering out commentary, errata, news articles, etc. which otherwise turn up and can clog your feed. - Use
NOT review[pt]to filter out reviews; see here. - Use
last X years[dp]to filter by recency; see here.
Issues? Questions? Suggestions?
Please add any issues you find to the Github issue tracker here. PRs are also welcome. I’ll try to address them as soon as I can. You can also email me at hello [at] hershbhargava.com with any questions or suggestions.
Footnotes
-
You can easily change the
ORbetween author and keyword/journal toANDif you want to only see papers that match both criteria. ↩ -
Note that the RSS link autogeneration feature of Pubmed-Sieve works using a headless browser (Selenium), since there is no API to create RSS feeds. This may break if Pubmed changes their UI. You can manually generate the RSS link by going to the PubMed search page, running a search, and clicking the “Create RSS” link below the search box. ↩